YAML simulation specification
Warning
This reference guide assumes that the reader knows the basics of YAML.
A good resource to learn it quickly is Learn X in Y minutes where X = YAML
Reading this document
The key words MUST, MUST NOT, REQUIRED, SHALL, SHALL NOT, SHOULD, SHOULD NOT,
RECOMMENDED, MAY, and OPTIONAL in this document are to be interpreted as described in
RFC 2119.
Simulation document structure
The document MUST be YAML map.
The map MUST contain all the mandatory Alchemist keys,
MAY contain any subset of the optional Alchemist keys,
MAY contain any key whose name begins with underscore (_),
and MUST NOT contain any other key.
The sets of valid cobinations of mandatory and optional keys for each section of the document is specified
in form of Kotlin code as follows:
Types of entries
| Type |
Description |
| Any |
Any YAML type |
| Int |
YAML integer number, or other type that can be parsed into an integer |
| List |
Any YAML List |
| Map |
Any YAML Map |
| MultiSpec |
A list of Spec. A Map matches a MultiSpec if it matches one and only one of its Spec. |
| Number |
YAML number |
| Spec |
Pair of lists of strings. The first list contains mandatory keys, the second optional keys. A map matches a Spec if it contains all its mandatory keys, any of the optional keys, and no other key |
| SpecMap |
A YAML Map matching a MultiSpec |
| String |
YAML String |
| Traversable |
One of: A SpecMap, a List of Traversable, a Map of Traversable |
Arbitrary class loading system
Type: SpecMap
Alchemist is able to load arbitrary types conforming to the expected interface
(or Scala trait).
The expected type depends on where the class is requested.
This section describes how the system works independently of the specific target type.
(Multi)Spec
| Mandatory keys |
Optional keys |
type |
parameters |
type
Type: String
The name of an instanceable class compatible with the expected interface.
It can be either the qualified name or a simple name,
in the latter case the class SHOULD be located in the same package where the
default alchemist implementations of the same interface live.
If a name includes a ., it is interpreted as a fully qualified name.
Otherwise, it is interpreted as a simple name.
Provided types SHOULD NOT be located in the default package.
For instance, if the expected type is an Action and the concrete type FooAction,
FooAction SHOULD be located into package it.unibo.alchemist.model.actions.
parameters
Type: List or Map
The list of parameters the constructor of type should be passed.
Alchemist automatically provides contextual information to the constructors:
for instance, if an Action is being built,
the loading system is aware of the current
RandomGenerator,
Incarnation,
Environment,
Deployment,
Node, TimeDistribution, and
Reaction,
as the action requires all of them.
Consequently, all parameters of these types SHOULD NOT be manually specified
(on the other hand, the syntax is built to make it very difficult to do by mistake).
The constructor MAY fail if they are provided.
If a Map is provided instead of a List,
then the keys are interpred as the parameter names,
and their associated values as the corresponding parameter values.
Since Java 11 does not support named arguments,
this special invocation type is built around the Kotlin reflection,
thus, the concrete class whose constructor is being invoked MUST be written in Kotlin.
When using named arguments,
if at least one optional parameter is specified,
then all the previous optional parameters MUST be specified as well.
This limitation is due to the fact that Alchemist supports loading of JVM classes regardless of their origin language,
and, thus, the simulator must leverage constructor overloading to emulate optional parameters.
In the case of Kotlin classes, because of the way
@JvmOverloads
works, only a (reasonable) subset of all possible overloads gets generated, and they differ by parameter count.
Instantiation is delegated to the Java Implicit Reflective Factory.
Examples
- Construction of a
Point
- Construction of variables with named parameters
Click to show / hide code
incarnation: sapere
network-model: { type: ConnectWithinDistance, parameters: [0.5] }
deployments:
type: GraphStreamDeployment
parameters:
nodeCount: 1000
offsetX: -10
offsetY: 10
zoom: 100
layoutQuality: 0.2
generatorName: Lobster
createLinks: true
parameters: [5, 15]
Counter-examples
- The following simulation fails on loading, as
BidimensionalGaussianLayer
has the first and last parameters marked as optional:
in order to provide the latter, the designer must also provide the former.
Click to show / hide code
incarnation: protelis
network-model:
type: ConnectWithinDistance
parameters: [6]
deployments:
type: Rectangle
parameters:
[1, 0, 0, 100, 100]
contents:
molecule: grain
concentration: 5
layers:
- molecule: layer
type: BidimensionalGaussianLayer
parameters:
centerX: 30
centerY: 30
norm: 30
sigmaX: 15
Document root
Type: SpecMap
The document contents at the root of the file.
Contains all the information required to buld a
Loader,
which, in turn, is able to spawn
Simulations
through a
Launcher.
(Multi)Spec
| Mandatory keys |
Optional keys |
incarnation |
deployments, environment, export, layers, launcher, network-model, remote-dependencies, seeds, terminate, variables |
Examples
- Minimal Biochemistry specification
- Minimal Protelis specification
- Minimal SAPERE specification
incarnation
Type: String
Valid incarnation types in the full distribution:
biochemistryprotelissaperescafi
Examples
- Minimal Biochemistry specification
- Minimal Protelis specification
- Minimal SAPERE specification
action
Builds an Action
using the arbitrary class loading system.
condition
Builds a Condition
using the arbitrary class loading system.
deployments
Type: Traversable
Traversable of deployment
deployment
Type: SpecMap
Definition of the positions of a set of nodes.
Builds a
Deployment
using the same syntax of arbitrary class loading system,
with additional keys.
(Multi)Spec
| Mandatory keys |
Optional keys |
type |
parameters, contents, nodes, programs |
Examples
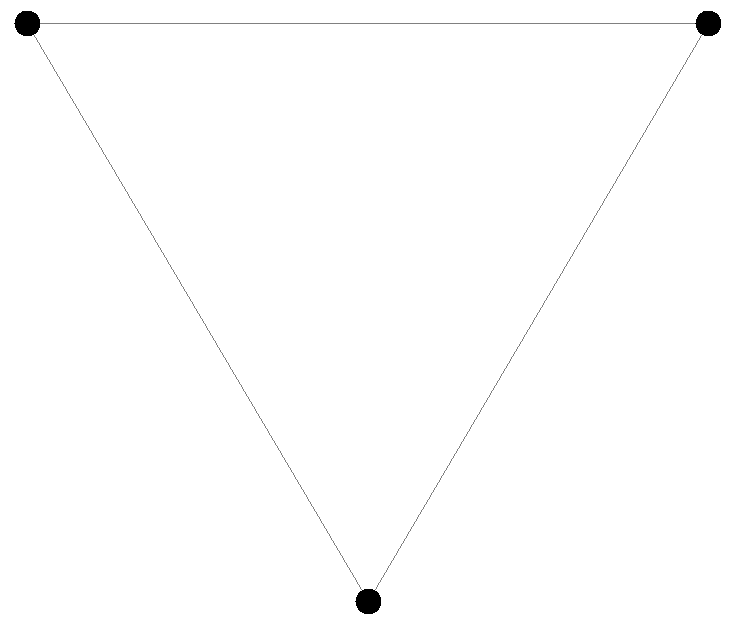
- Deployment of a single node in a point
- Deployment of three nodes
Click to show / hide code
incarnation: sapere # The incarnation is always mandatory
network-model:
type: ConnectWithinDistance # Loads a class with this name implementing LinkingRule
parameters: [2] # Connection radius (parameter of a ConnectWithinDistance's constructor)
deployments:
- type: Point # Loads a class with this name implementing Deployment
parameters: [0, 0] # Coordinates
- { type: Point, parameters: [0.5, 0.85] }
- { type: Point, parameters: [-0.5, 0.85] }
- Deployment of three nodes, but nesting the traversable
Click to show / hide code
incarnation: protelis
deployments:
the_bottom_point:
type: Point
parameters: [0, 0]
the_top_points:
- type: Point
parameters: [0.5, 0.85]
- type: Point
parameters: [-0.5, 0.85]
- Deployment of three nodes through
SpecificPositions.
Grid centered in (0, 0), with nodes distanced of 0.25 both horizontally and vertically.
- Irregular
Grid centered in (0, 0), with nodes distanced of 0.25 both horizontally and vertically, randomly perturbed of (±0.1 distance units).
- Nodes located randomly inside a
Circle
- Nodes located randomly inside a
Rectangle
- Nodes located randomly inside a
Polygon delimiting the Venice Lagoon
Click to show / hide code
incarnation: sapere
environment: { type: OSMEnvironment }
network-model: { type: ConnectWithinDistance, parameters: [1000] }
_venice_lagoon: &lagoon
[[45.2038121, 12.2504425], [45.2207426, 12.2641754], [45.2381516, 12.2806549],
[45.2570053, 12.2895813], [45.276336, 12.2957611], [45.3029049, 12.2991943],
[45.3212544, 12.3046875], [45.331875, 12.3040009], [45.3453893, 12.3040009],
[45.3502151, 12.3156738], [45.3622776, 12.3232269], [45.3719259, 12.3300934],
[45.3830193, 12.3348999], [45.395557, 12.3445129], [45.3998964, 12.3300934],
[45.4018249, 12.3136139], [45.4105023, 12.3122406], [45.4167685, 12.311554],
[45.4278531, 12.3012543], [45.4408627, 12.2902679], [45.4355628, 12.2772217],
[45.4206242, 12.2703552], [45.3994143, 12.2744751], [45.3738553, 12.2676086],
[45.3579354, 12.2614288], [45.3429763, 12.2497559], [45.3198059, 12.2408295],
[45.2975921, 12.2346497], [45.2802014, 12.2408295], [45.257972, 12.233963],
[45.2038121, 12.2504425]]
deployments:
type: Polygon
parameters: [500, *lagoon]
programs:
- time-distribution: 10
type: Event
actions: { type: BrownianMove, parameters: [0.0005]}
deployment.type
Same as type
deployment.parameters
Same as parameters
deployment.contents
Type: Traversable of content
deployment.nodes
Type: SpecMap
Forces the type of Node, building concrete types through
the arbitrary class loading system.
If left unspecified, nodes get created through
Incarnation.createNode.
Examples
- Creation of heterogeneous pedestrians
Click to show / hide code
incarnation: protelis
environment:
type: ContinuousPhysics2DEnvironment
seeds:
scenario: 0
simulation: 1
deployments:
- type: Circle
parameters: [50, 0, 0, 20]
properties:
- type: Human
parameters: ["elderly", "female"]
- type: HeterogeneousPedestrian
- type: Perceptive2D
- type: CircularArea
- type: Social
- type: Circle
parameters: [50, 0, 0, 20]
properties:
- type: Human
parameters: ["child", "male"]
- type: HeterogeneousPedestrian
- type: Perceptive2D
- type: CircularArea
- type: Social
deployment.properties
Type: Traversable of property
deployment.programs
Type: Traversable of program
content
Type: SpecMap
Definition of the contents (Molecules and Concentrations) of a group of nodes.
(Multi)Spec
| Mandatory keys |
Optional keys |
molecule, concentration |
in |
Examples
- Three molecules injected into all nodes deployed in the scenario
Click to show / hide code
incarnation: protelis
network-model:
type: ConnectWithinDistance
parameters: [7]
_gradient: &gradient
- time-distribution: 1
type: Event
actions:
- type: RunProtelisProgram
parameters: [distanceTo, 1.01]
- program: send
deployments:
- type: Point
parameters: [0,0]
contents:
- molecule: source
concentration: false
- molecule: enabled
concentration: true
- molecule: data
concentration: 1 / 0
programs: *gradient
terminate:
type: StepCount
parameters: 5000
- Injection of a molecule only in those nodes located inside a
Rectangle
Click to show / hide code
incarnation: sapere
network-model: { type: ConnectWithinDistance, parameters: [0.5] }
deployments:
type: Grid
parameters: [-5, -5, 5, 5, 0.25, 0.25, 0.1, 0.1] # A perturbed grid of devices
contents:
- molecule: "{hit, 0}" # Everywhere, no one has been hit
- in: { type: Rectangle, parameters: [-0.5, -0.5, 1, 1] } # Inside this shape...
molecule: ball # ...every node has a ball
programs:
- time-distribution: 1 # This is a frequency, time distribution type is left to the incarnation
# 'program' specs are passed down to the incarnation for being interpreted as reactions
program: "{ball} {hit, N} --> {hit, N + 1} {launching}" # If hit, count the hit
- program: "{launching} --> +{ball}" # As soon as possible, throw the ball to a neighbor
content.molecule
Type: String or SpecMap
The name of the molecule to be injected.
If a String is provided, then it is created via Incarnation.createMolecule.
Otherwise, the arbitrary class loading system SHOULD be used.
content.concentration
Type: String
The concentration of the molecule to be injected.
If a String is provided, then it is created via Incarnation.createConcentration.
Otherwise, the arbitrary class loading system SHOULD be used.
content.in
Type: Traversable of shapeFilter
property
Type: SpecMap
(Multi)Spec
| Mandatory keys |
Optional keys |
type |
parameters, in |
property.type
Same as type
property.parameters
Same as parameters
property.in
Type: Traversable of shapeFilter
environment
Type: SpecMap
Builds an Environment
using the same syntax of arbitrary class loading system.
If left unspecified, defaults to a bidimensional Euclidean manifold:
Continuous2DEnvironment.
Type: SpecMap
Examples
- Default environment, omitted specification
- Explicitly builds a
Continuous2DEnvironment solely with the contextual parameters
- Explicitly builds a
Continuous2DEnvironment using the qualified type name using only the contextual parameters
Click to show / hide code
incarnation: protelis
environment:
type: it.unibo.alchemist.model.environments.Continuous2DEnvironment
- Explicitly builds a
Continuous2DEnvironment explicitly specifying that no parameters but the contextual ones should be used
export
Type: Traversable of exporter
exporter
Type: SpecMap
Definition of the contents (Molecules and Concentrations) of a group of nodes.
(Multi)Spec
| Mandatory keys |
Optional keys |
type, data |
parameters |
exporter.type
Same as type
exporter.data
Type: Traversable of extractor
exporter.parameters
Same as parameters
Type: String or SpecMap
The only supported String is "time".
Otherwise, a SpecMap MUST be provided.
Creates instances of Extractor.
(Multi)Spec
| Mandatory keys |
Optional keys |
type |
parameters |
molecule |
property, aggregators, value-filter |
Same as type
Same as parameters
Type: String
Name of a Molecule to be read from nodes and exported.
The String is passed down to Incarnation.createMolecule.
The created molecule is read from every node.
Type: String
Name of a property to be extracted from the selected Molecule.
The Molecule and the String are passed down to Incarnation.getProperty.
The obtained value is added to the exports.
Type: String or List of Strings
Name of any valid
UnivariateStatistic,
case insensitive.
All those provided with Apache Commons Math are available by default.
New statistics can be defined,
they get loaded transparently as far as their package matches the one of Apache Commons Math.
If the aggregators are specified, only one value per aggregator gets exported,
instead of one value for each node.
Type: String or SpecMap
Builds a ExportFilter,
to be applied to raw data before being processed by the aggregators(#extractoraggregators),
if present.
If a String is provided, then it is used to load a policy from CommonFilters.
Otherwise, the arbitrary class loading system MUST be used.
| Mandatory keys |
Optional keys |
type |
parameters |
Same as type
Same as parameters
launcher
Type: SpecMap
Builds a Launcher
using the arbitrary class loading system.
If unspecified, defaults to DefaultLauncher,
which runs the default simulation,
unless the variable set on which the batch should be executed is specified.
Customizing the launcher can be useful for implementing custom batch execution strategies,
or “simulations of simulations”,
if the process requires multiple simulation “stages”
(e.g., running a batch to train a neural network, then running another batch to test it).
layer
Type: SpecMap
Builds a Layer
using the arbitrary class loading system.
Examples
- Creation of two
Layers
Click to show / hide code
incarnation: sapere
environment:
type: Continuous2DEnvironment
parameters: []
layers:
- type: StepLayer
parameters: [2, 2, 100, 0]
molecule: A
- type: StepLayer
parameters: [-2, -2, 0, 100]
molecule: B
- Creation of two
BidimensionalGaussianLayers:
Click to show / hide code
incarnation: protelis
_danger: &danger
danger
_target: &target
targe
environment:
type: ContinuousPhysics2DEnvironment
layers:
- type: BidimensionalGaussianLayer
molecule: *danger
parameters: [80, 0, 100, 20]
- type: BidimensionalGaussianLayer
molecule: *target
parameters: [-50, 0, 10, 50]
seeds:
scenario: 0
simulation: 1
_reactions: &behavior
- time-distribution:
type: DiracComb
parameters: [2.0]
type: BlendedSteering
actions:
- type: CognitiveAgentAvoidLayer
parameters: [*danger]
- type: CognitiveAgentFollowLayer
parameters: [*target]
conditions:
- type: WantToEscape
- time-distribution:
type: DiracComb
parameters: [0.5]
type: CognitiveBehavior
actions:
- type: HeadTowardRandomDirection
_nodes: &nodes
properties:
- type: Human
parameters: [ "adult", "male" ]
- type: CognitivePedestrian
- type: Cognitive2D
parameters: [ *danger ]
- type: Perceptive2D
- type: CircularArea
programs: *behavior
deployments:
- type: Circle
parameters: [25, 0, 0, 8]
<<: *nodes
- type: Circle
parameters: [75, 60, 0, 10]
<<: *nodes
layers
Type: Traversable of layer
Examples
- Creation of two
Layers
Click to show / hide code
incarnation: sapere
environment:
type: Continuous2DEnvironment
parameters: []
layers:
- type: StepLayer
parameters: [2, 2, 100, 0]
molecule: A
- type: StepLayer
parameters: [-2, -2, 0, 100]
molecule: B
- Creation of two
BidimensionalGaussianLayers:
Click to show / hide code
incarnation: protelis
_danger: &danger
danger
_target: &target
targe
environment:
type: ContinuousPhysics2DEnvironment
layers:
- type: BidimensionalGaussianLayer
molecule: *danger
parameters: [80, 0, 100, 20]
- type: BidimensionalGaussianLayer
molecule: *target
parameters: [-50, 0, 10, 50]
seeds:
scenario: 0
simulation: 1
_reactions: &behavior
- time-distribution:
type: DiracComb
parameters: [2.0]
type: BlendedSteering
actions:
- type: CognitiveAgentAvoidLayer
parameters: [*danger]
- type: CognitiveAgentFollowLayer
parameters: [*target]
conditions:
- type: WantToEscape
- time-distribution:
type: DiracComb
parameters: [0.5]
type: CognitiveBehavior
actions:
- type: HeadTowardRandomDirection
_nodes: &nodes
properties:
- type: Human
parameters: [ "adult", "male" ]
- type: CognitivePedestrian
- type: Cognitive2D
parameters: [ *danger ]
- type: Perceptive2D
- type: CircularArea
programs: *behavior
deployments:
- type: Circle
parameters: [25, 0, 0, 8]
<<: *nodes
- type: Circle
parameters: [75, 60, 0, 10]
<<: *nodes
monitor
Type: SpecMap
Builds a OutputMonitor
using the arbitrary class loading system.
Examples
- Creation of one
OutputMonitors
Click to show / hide code
incarnation: sapere
environment:
type: Continuous2DEnvironment
global-programs:
- time-distribution:
type: DiracComb
parameters: [1.0]
type: GlobalTestReaction
monitors:
- type: another.location.SimpleMonitor
monitors
Type: Traversable of monitor
Examples
- Creation of one
OutputMonitors
Click to show / hide code
incarnation: sapere
environment:
type: Continuous2DEnvironment
global-programs:
- time-distribution:
type: DiracComb
parameters: [1.0]
type: GlobalTestReaction
monitors:
- type: another.location.SimpleMonitor
network-model
Type: SpecMap
Builds a LinkingRule
using the arbitrary class loading system.
If unspecified, defaults to NoLinks,
and no node will have any neighbor.
Examples
- Nodes connected when closer than some range
Click to show / hide code
incarnation: sapere # The incarnation is always mandatory
network-model:
type: ConnectWithinDistance # Loads a class with this name implementing LinkingRule
parameters: [2] # Connection radius (parameter of a ConnectWithinDistance's constructor)
deployments:
- type: Point # Loads a class with this name implementing Deployment
parameters: [0, 0] # Coordinates
- { type: Point, parameters: [0.5, 0.85] }
- { type: Point, parameters: [-0.5, 0.85] }
program
Type: SpecMap
Definition of the contents (Molecules and Concentrations) of a group of nodes.
(Multi)Spec
| Mandatory keys |
Optional keys |
type |
parameters, conditions, time-distribution actions |
program |
time-distribution |
program.type
Same as type
program.program
Type: String
Passed to Incarnation.createReaction to be interepreted and
program.in
Type: Traversable of shapeFilter
program.actions
Type: Traversable of action
program.conditions
Type: Traversable of condition
program.parameters
Same as parameters
program.time-distribution
Type: String or SpecMap
Builds a TimeDistribution.
If a String is provided, then it is created via Incarnation.createTimeDistribution.
Otherwise, the arbitrary class loading system SHOULD be used.
remote-dependencies
shapeFilter
Type: SpecMap
Builds a PositionBasedFilter
using the arbitrary class loading system.
Examples
- Injection of a molecule only in those nodes located inside a
Rectangle
Click to show / hide code
incarnation: sapere
network-model: { type: ConnectWithinDistance, parameters: [0.5] }
deployments:
type: Grid
parameters: [-5, -5, 5, 5, 0.25, 0.25, 0.1, 0.1] # A perturbed grid of devices
contents:
- molecule: "{hit, 0}" # Everywhere, no one has been hit
- in: { type: Rectangle, parameters: [-0.5, -0.5, 1, 1] } # Inside this shape...
molecule: ball # ...every node has a ball
programs:
- time-distribution: 1 # This is a frequency, time distribution type is left to the incarnation
# 'program' specs are passed down to the incarnation for being interpreted as reactions
program: "{ball} {hit, N} --> {hit, N + 1} {launching}" # If hit, count the hit
- program: "{launching} --> +{ball}" # As soon as possible, throw the ball to a neighbor
seeds
Type: SpecMap
Selection of the seed for the
RandomGenerators.
(Multi)Spec
| Mandatory keys |
Optional keys |
|
scenario, simulation |
seeds.scenario
Type: Int
Selection of the seed for the
RandomGenerator
controlling the position of random displacements.
seeds.simulation
Type: Int
Selection of the seed for the
RandomGenerator
controlling the evolution of the events of the simulation.
terminate
Type: Traversable of terminator
terminator
Type: SpecMap
Builds a Predicate
using the arbitrary class loading system.
Examples
- Termination after some time
Click to show / hide code
incarnation: protelis
network-model:
type: ConnectWithinDistance
parameters: [30]
deployments:
- type: Rectangle
parameters: [100, 62, 15, 95, 200]
contents:
- molecule: "source"
concentration: true
in:
type: Circle
parameters: [107.96487911806524, 102.49167432603535, 10]
programs:
- time-distribution: 1 # This is a frequency
program: >
import protelis:coord:spreading
distanceTo(self.getDeviceUID().getId() == 0)
- program: send
terminate:
- type: AfterTime
parameters: 1
Click to show / hide code
incarnation: protelis
variables:
zoom: &zoom
formula: 0.1
image_name: { formula: "'chiaravalle.png'" }
image_path: &image_path
language: kotlin
formula: >
import java.io.File
File("../..").walkTopDown().find { image_name in it.name }?.absolutePath ?: image_name
walking_speed: &walk-speed { default: 1.4, min: 1, max: 2, step: 0.1 }
seed: &seed { default: 0, min: 0, max: 99, step: 1 }
scenario_seed: &scenario_seed { formula: (seed + 31) * seed }
people_count: &people_count
type: GeometricVariable
parameters: [300, 50, 500, 9]
seeds: { simulation: *seed, scenario: *scenario_seed}
export:
- type: CSVExporter
parameters:
fileNameRoot: "00-testing_csv_export"
data:
- time
- molecule: "default_module:default_program"
aggregators: [ mean, max, min, variance, median ]
value-filter: onlyfinite
- type: CSVExporter
parameters:
fileNameRoot: "fixed-decimals"
data:
- type: Time
parameters:
precision: 2
- molecule: "default_module:default_program"
property: "self.nextRandomDouble()"
precision: 2
aggregators: [ mean, max, min, variance, median ]
value-filter: onlyfinite
environment: { type: ImageEnvironment, parameters: [*image_path, *zoom] }
network-model: { type: ObstaclesBreakConnection, parameters: [5] }
deployments:
type: Rectangle
parameters: [*people_count, 62, 15, 95, 200]
programs:
- time-distribution: 1
program: >
import protelis:coord:spreading
let source = [110, 325]
let vector = self.getCoordinates() - source
let distance = hypot(vector.get(0), vector.get(1))
distanceTo(distance < 50)
- program: send
- { type: Event, time-distribution: 1, actions: { type: LevyWalk, parameters: [*walk-speed] } }
terminate:
- type: AfterTime
parameters: 10
Click to show / hide code
incarnation: protelis
variables:
zoom: &zoom
formula: 0.1
image_name: { formula: "'chiaravalle.png'" }
image_path: &image_path
language: kotlin
formula: >
import java.io.File
File("../..").walkTopDown().find { image_name in it.name }?.absolutePath ?: image_name
walking_speed: &walk-speed { default: 1.4, min: 1, max: 2, step: 0.1 }
seed: &seed { default: 0, min: 0, max: 99, step: 1 }
scenario_seed: &scenario_seed { formula: (seed + 31) * seed }
people_count: &people_count
type: GeometricVariable
parameters: [10, 50, 500, 9]
seeds: { simulation: *seed, scenario: *scenario_seed}
export:
- type: MongoDBExporter
parameters:
uri: "mongodb://localhost:27017/"
dbName: "test"
interval: 2.0
data:
- time
- molecule: "default_module:default_program"
aggregators: [ mean, max, min, variance, median ]
value-filter: onlyfinite
environment: { type: ImageEnvironment, parameters: [*image_path, *zoom] }
network-model: { type: ObstaclesBreakConnection, parameters: [50] }
deployments:
type: Rectangle
parameters: [*people_count, 62, 15, 95, 200]
programs:
- time-distribution: 1
program: >
import protelis:coord:spreading
let source = [110, 325]
let vector = self.getCoordinates() - source
let distance = hypot(vector.get(0), vector.get(1))
distanceTo(distance < 50)
- program: send
- { type: Event, time-distribution: 1, actions: { type: LevyWalk, parameters: [*walk-speed] } }
terminate:
- type: AfterTime
parameters: 3
variable
Type: SpecMap
Definition of free
and dependent variables.
(Multi)Spec
| Mandatory keys |
Optional keys |
type |
parameters |
min, max, step, default |
|
formula |
language, timeout |
Variables can be created in three ways:
variable.type
Same as type
variable.parameters
Same as parameters
variable.default
Type: Number
Default value for a LinearVariable,
to be selected if the variable is not among those generating the batch.
variable.max
Type: Number
Maximum value for a LinearVariable
variable.min
Type: Number
Minimum value for a LinearVariable
variable.step
Type: Number
Size of the incremental step of a LinearVariable
Type: String
Code that can be interpreted by a JSR223Variable.
variable.language
Type: String
Language to be used by a JSR223Variable.
The language must be available in the classpath.
Groovy (default), Kotlin (kotlin or kts), and Scala (scala) are supported natively.
variable.timeout
Type: Int
Time in milliseconds after which the interpreter of the
JSR223Variable
is considered stuck or in livelock.
The interpreter gets interrupted and the simulation loading fails to prevent unresponsive simulations.
Defaults to 1000ms.
variables
Type: Traversable of variable
Biochemistry Incarnation
The Biochemistry DSL
Biochemistry programs are written in a and human-readable syntax.
Valid programs can be written directly into
Those simple reactions can be fed directly as program in the YAML file.
Reactions
A reaction rule can be set using the symbol --> according to chemistry equations,
and placing both the molecules and the actions inside two square brackets
(ex. [OH], [H2O], [BrownianMove(0.1)])
The following line represents a basic chemical reaction that happens inside a cell:
[H] + [OH] --> [H2O]
However, reactions can also take place outside cells.
Biological cells, indeed, can swap molecules with its neighbour or the surrounding environment,
and this is possible in Alchemist too, using the keywords:
in cell, in neighbour and in env.
The reaction [A in env] --> [A in cell] moves the molecule A from the environement inside the cell.
If the location is not explicit, it is assumed the molecule to be inside the cell.
Junctions
A junction can be created just with a neighbor of the programmed cell.
The way to create it is with the syntax [X] + [Y in neighbor] --> [junction X-Y],
which means that when this reaction happens a junction using the molecule X
from the cell and the molecule Y from the neighbor will be created.
The junction can also be destroyed using the syntax [junction X-Y] --> [],
causing the reintroduction of the molecule X inside the cell and the molecule Y
inside the neighbor.
Also, the junction will be automatically removed if, because of their movement,
the cells will stop being in a neighborhood.
Custom Conditions
Any custom condition must be placed after the reaction products following an if clause.
For example, to create a molecule if the cell has at least three neighbor you would write:
[] --> [X] if NumberOfNeighborsGreaterThan(5)
Movement
A movement can be performed in the same way of a reaction, using the function as it is a product of the reaction itself.
This program constantly moves a cell without any other condition:
[] --> [BrownianMove(0.1)]
Collisions
The Biochemistry Incarnation supports cell collisions and deformations too.
In order to do that, however, the environment must feature appropriate support, as for instance
BioRect2DEnvironmentNoOverlap.
The cells must support deformation as well, as, for instance, a node with the
CircularDeformableCell
property.
The minimum radius of the cell is so that min-radius = rigidity * max-radius
and the two parameters are used to compute collisions and impacts between the cells.
Command Line interface
The CLI Interface
Alchemist utilizes a CLI interface to run a simulation.
A minimal launch looks like this:
run --simulationFile simulation.yml
Where the options are
run - Tells that Alchemist simulation is to be runned--simulationFile - Indicates the resource or path to the resource for the simulation configuration file
Logging Verbosity
Unless specifies, Alchemist logs with the warn logging level by default. Logging level tells
how verbose and throrough the outputted logs are.
Alchemist has the following logging levels avaialble (from less to most verbose):
- off
- debug
- info
- warn
- error
- all
In order to specify verbosity, the --verbosity option can be used:
run --simulationFile simulation.yml --verbosity error
Overriding Variables
Alchemist parses the configuration variables from the simulation configuration file.
In some cases it may be desirable to override some of the simulation file variables without
resorting to creating a new file. For such cases, --override option is available.
This options takes in input a valid yaml string representing the part of the configuration file to be overriden.
For example, given configuration file simulation.yml:
foo:
bar:
fizz: 42
buzz: some-string
And override with
run --simulationFile simulation.yml --override
foo:
bar:
buzz: 3
The resulting simulation file would be equivalent to
foo:
bar:
fizz: 42
buzz: 3
The overrides are arbitrary, types can be changed and new varibales introduced.
Launcher Configuration
Alchemist needs a Launcher class in order to run the simulation. Unless configured,
Alchemist will default to a launcher that runs the default configuration,
unless batch variables are explicitly provided: DefaultLauncher
If you would like to use another launcher class, you need to configure it in the simulation configuration file
as per the alchemist Arbitrary class loading system.
Here is an example of a headless simulation run with additional parameters:
cli options
run --simulationFile simulation.yml
simulation.yml
...
launcher:
type: DefaultLauncher
parameters:
parallelism: 4
variables: [ 1, 2, 3, 4 ]
...
Migrating From Legacy CLI
Here is a brief guide on how to re-map legacy CLI configuration options to the new
configuration flow.
-hl - Migrated to launcher configuration, use DefaultLauncher-var - Migrated to launcher configuration, used as parameters in supporting launchers-b - Migrated to launcher configuration-fxui - Migrated to launcher configuration-d - Migrated to launcher configuration-g - Migrated to launcher configuration, used as parameters in supporting launchers-h - Removed-s - Migrated to launcher configuration, used as parameters in supporting launchers-p - Migrated to launcher configuration, used as parameters in supporting launchers-t - Removed, use termination conditions instead (see examples below)-y - Removed, provide simulation file directly as program argument-w - Migrated to launcher configuration
Common Launch Configurations Snippets
SwingGUI Launch configuration
...
monitors:
type: SwingGUI
parameters:
graphics: /effects/some-effect.json
...
Terminate after 50 time units configuration
...
terminate:
- type: AfterTime
parameters: 50
...